Cell Signaling Technology is well known for their pathway diagrams. Inspired by the work of David Goodsell, Digizyme worked with CST to bring a new level of context to the pathways. Could we take the diagrams and show them in the crowded environment of a single cell, to highlight overlaps and connections between signals?


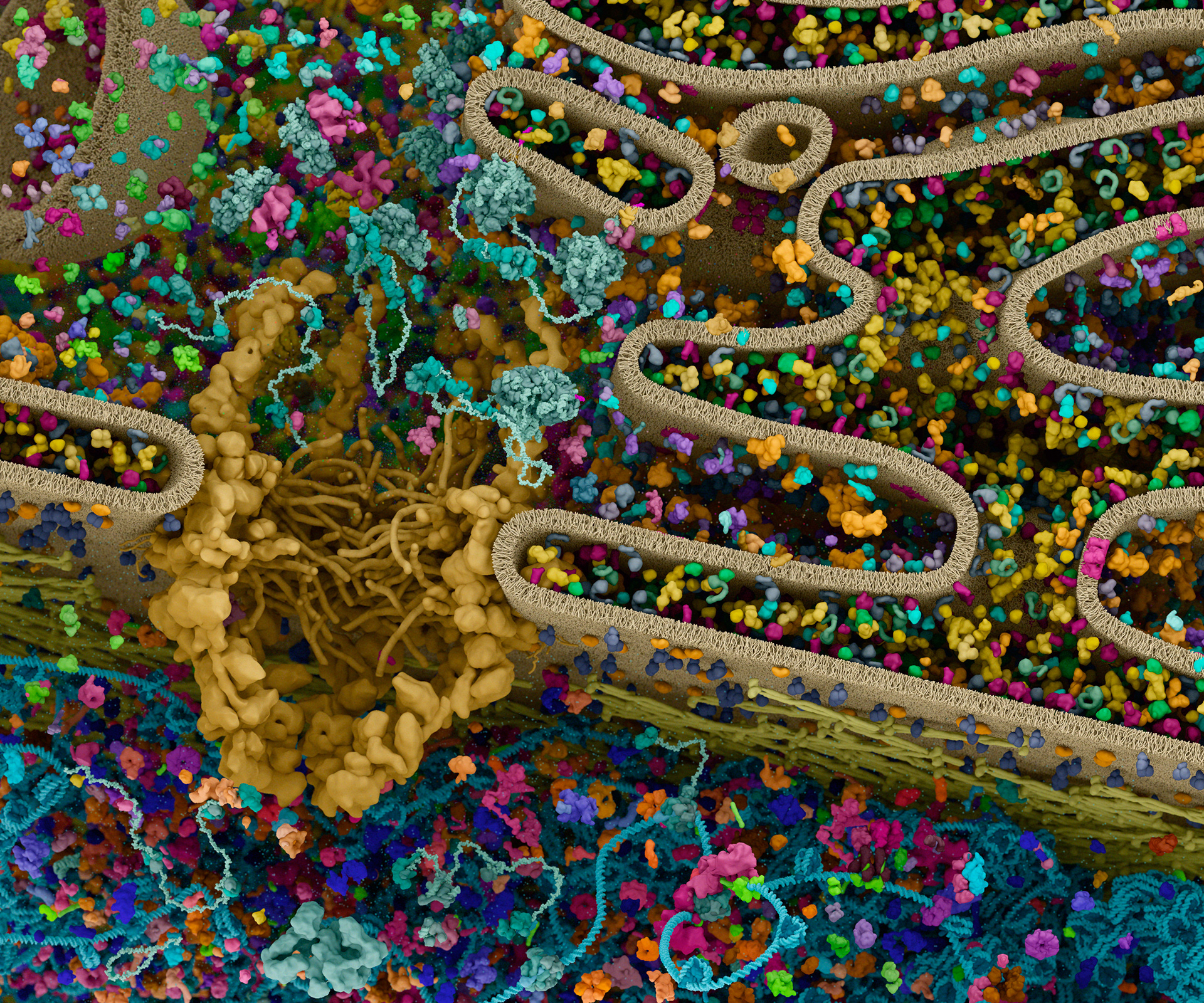
The challenges were immense. How to represent proteins with differing amounts of known structural data? How to distinguish hundreds of “hero” proteins from thousands of “background” elements? How could users interact with the images to filter out proteins or pathways of interest?
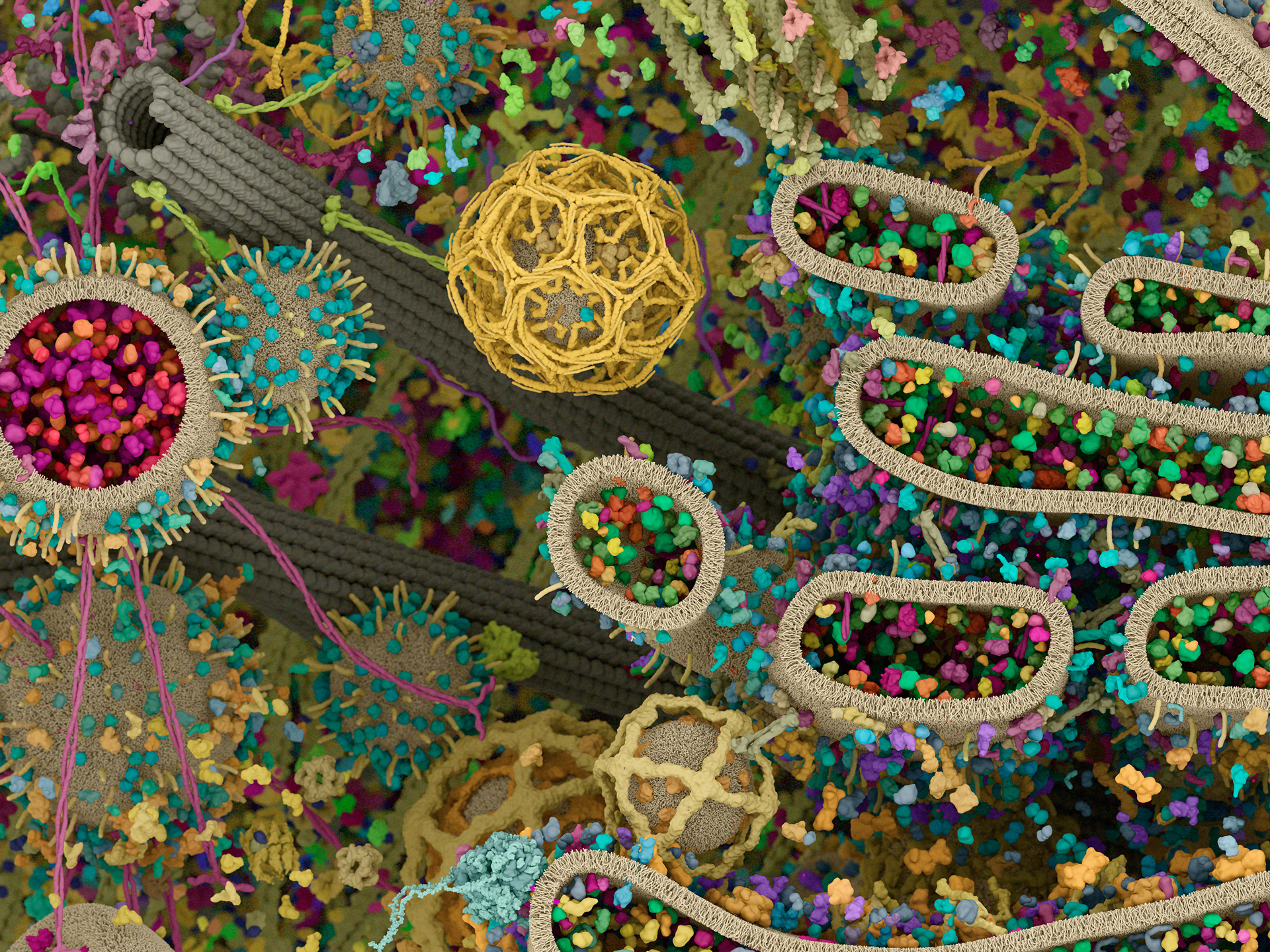
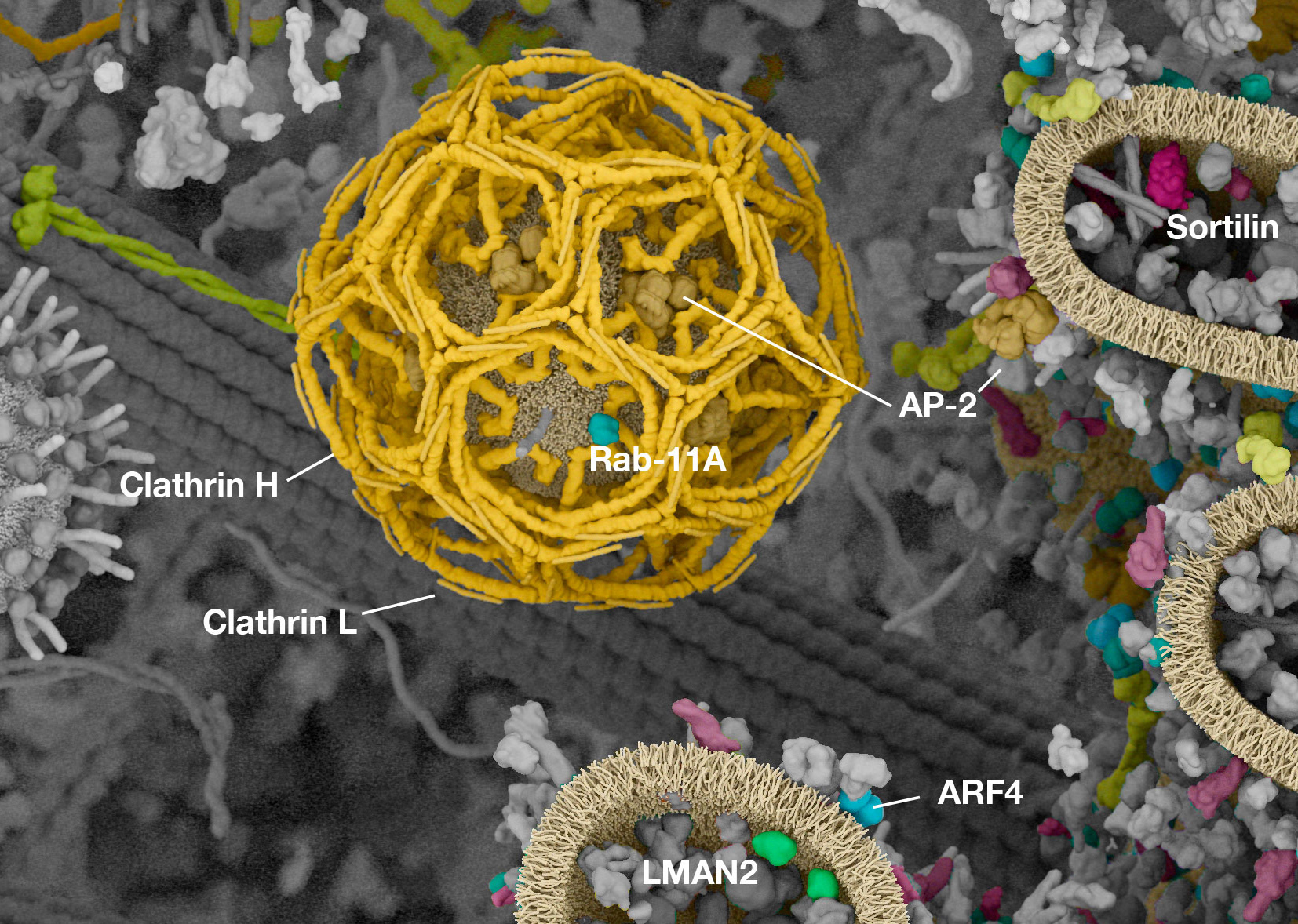
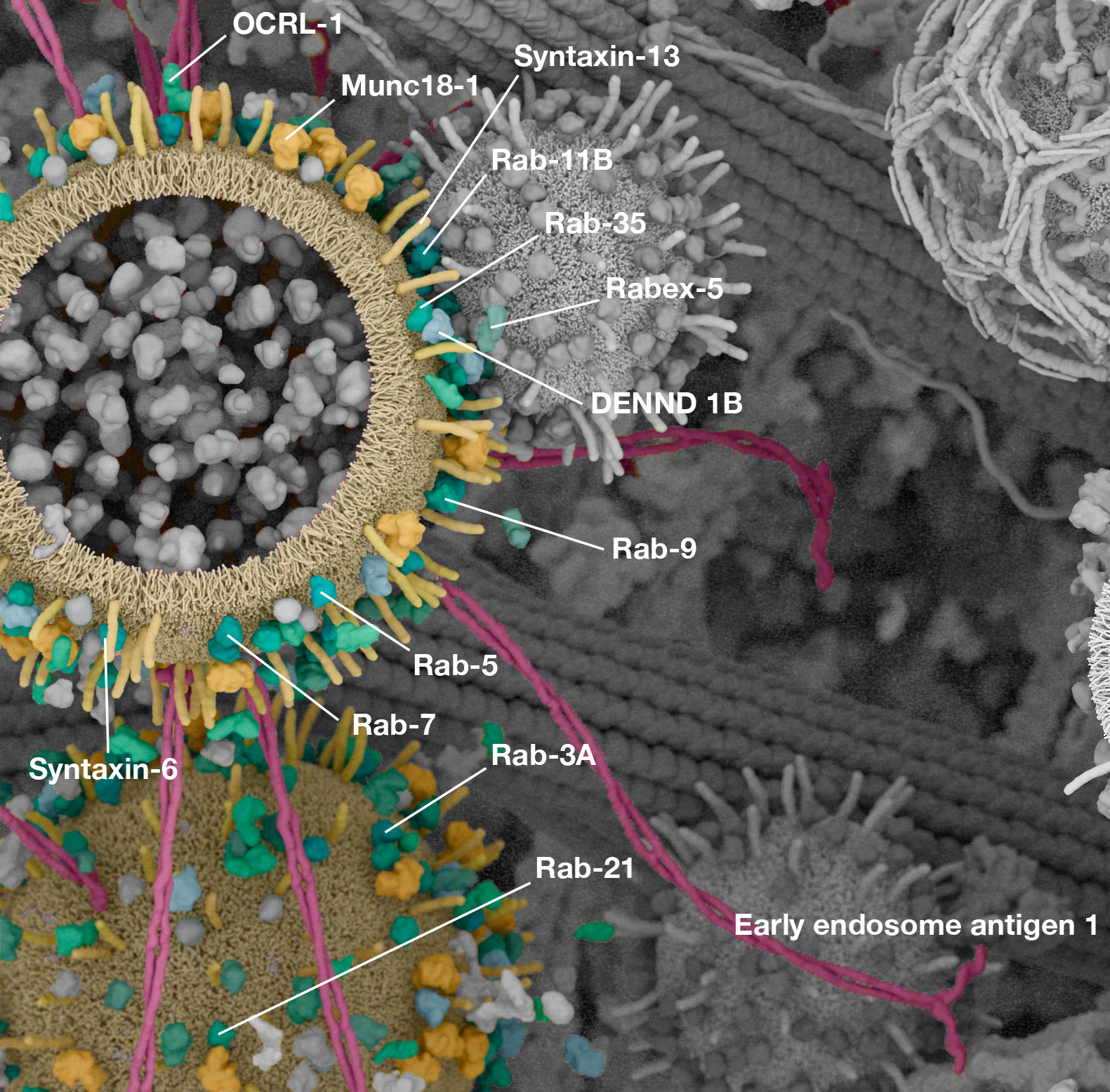
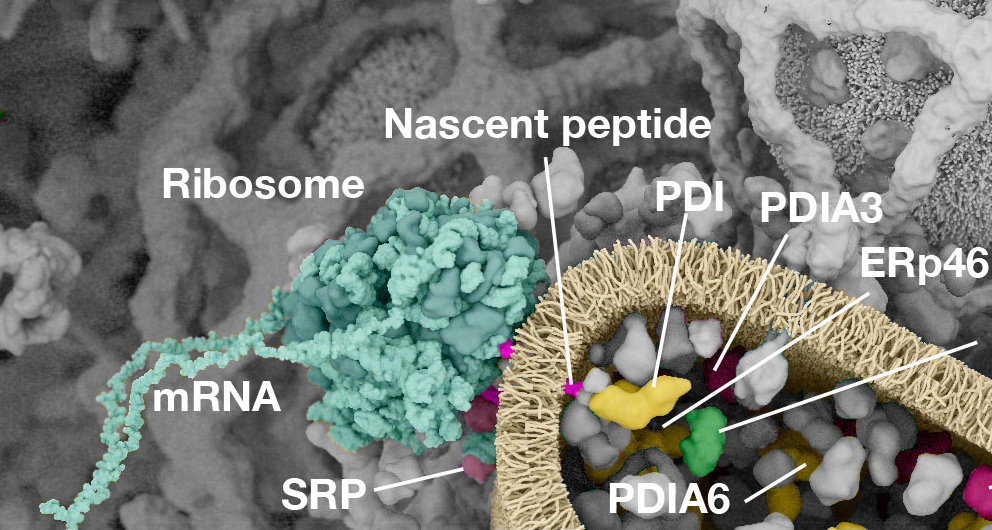
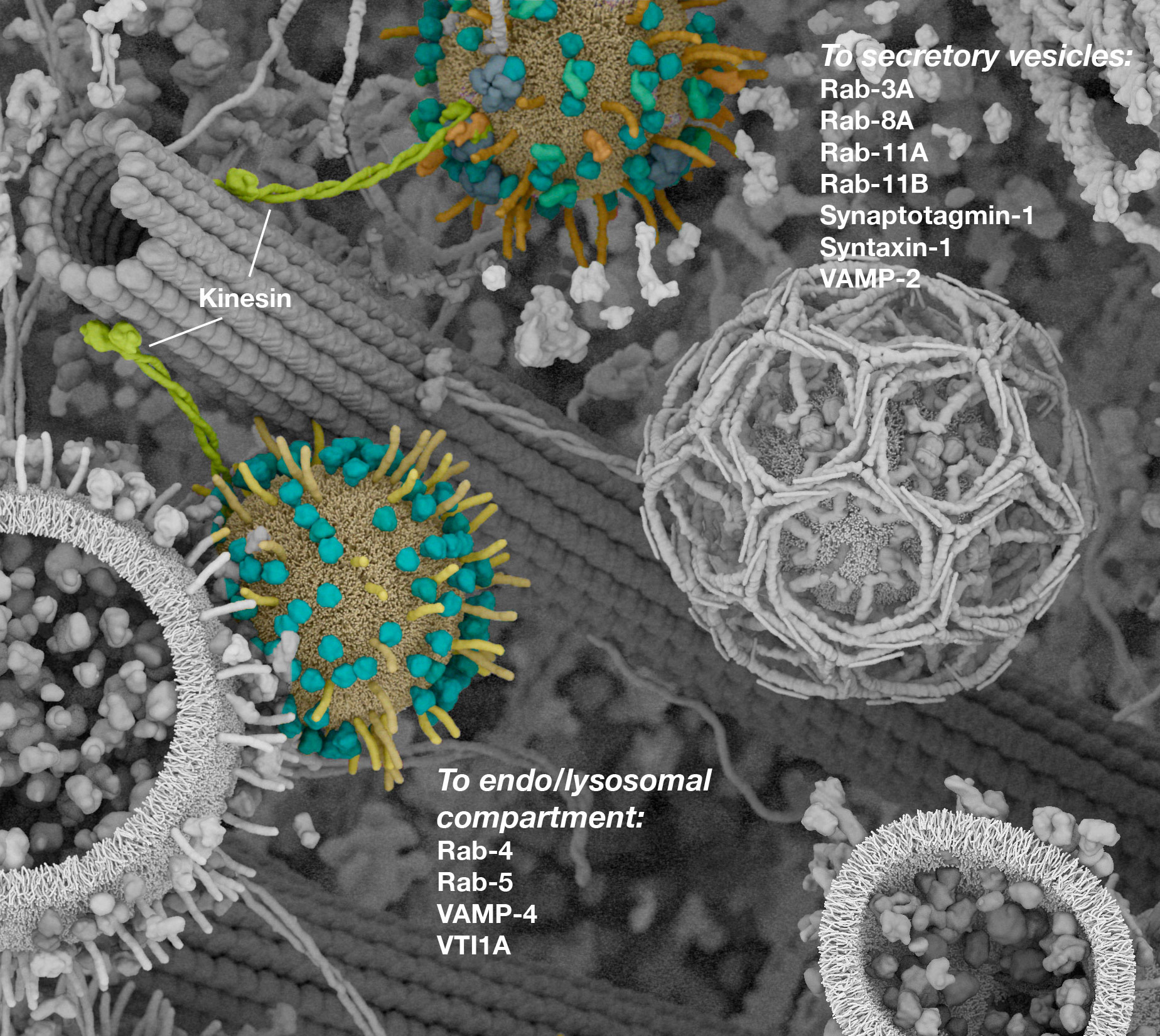
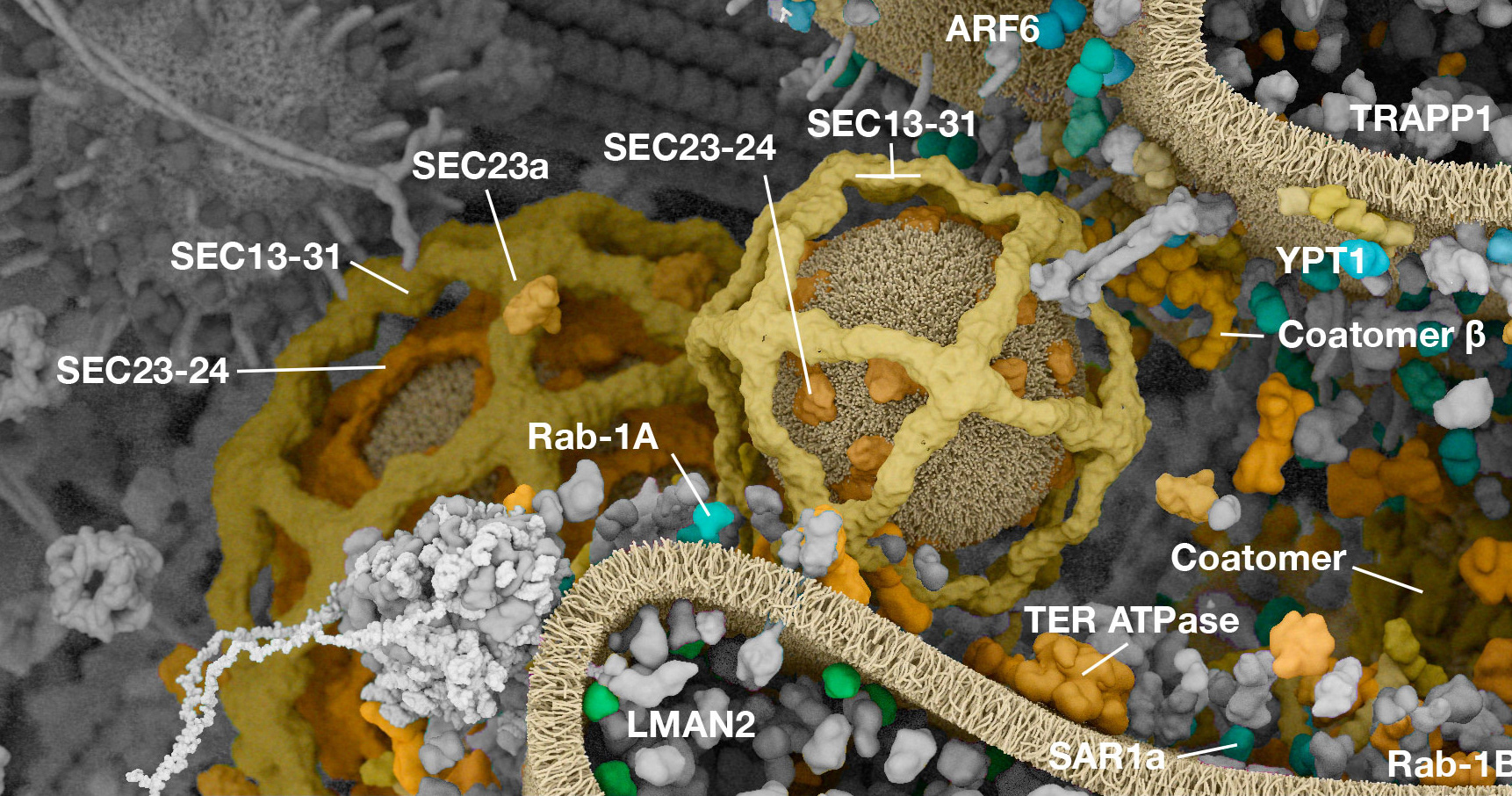
Working with (and eventually at) Digizyme over the course of six years, we produced six interactive landscapes that tile seamlessly into a large-scale poster. I encourage you to visit the interactive versions, which are available at web.archive.org. (The archived version is slower than the original, but appears to work as of 2025.)
client: Cell Signaling Technology
agency: Digizyme, Inc
roles: independent contractor, Angstrom Images, LLC (panels 1-3); Scientific Animator, Digizyme (panels 4-6, poster)
agency: Digizyme, Inc
roles: independent contractor, Angstrom Images, LLC (panels 1-3); Scientific Animator, Digizyme (panels 4-6, poster)
Concept, layout, art direction, interactive programming: Digizyme
Models, shading, renders, compositing, interactive pre-production: Evan Ingersoll
MODO, Molecular Maya, AfterEffects, Photoshop, Illustrator, NUKE
Models, shading, renders, compositing, interactive pre-production: Evan Ingersoll
MODO, Molecular Maya, AfterEffects, Photoshop, Illustrator, NUKE