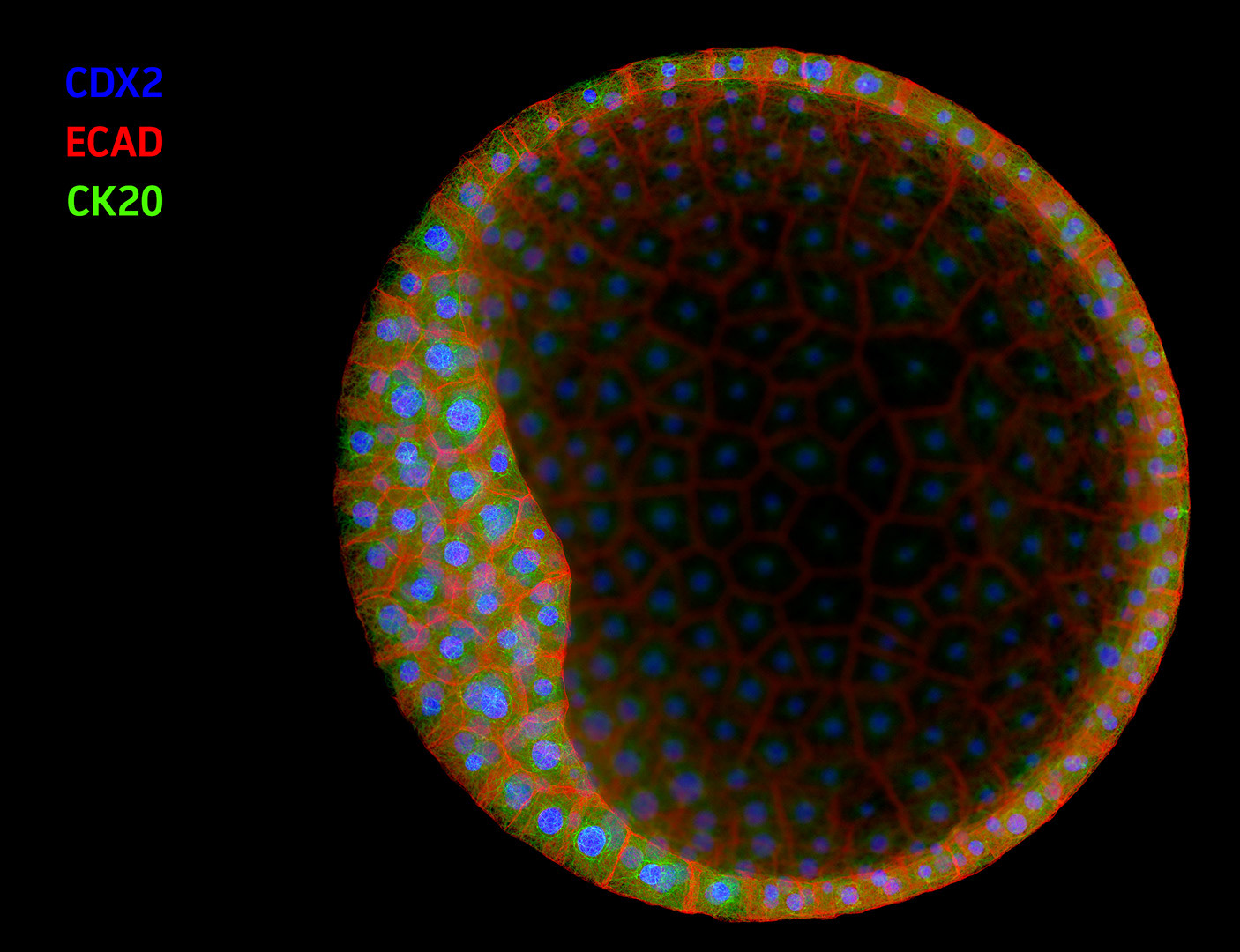
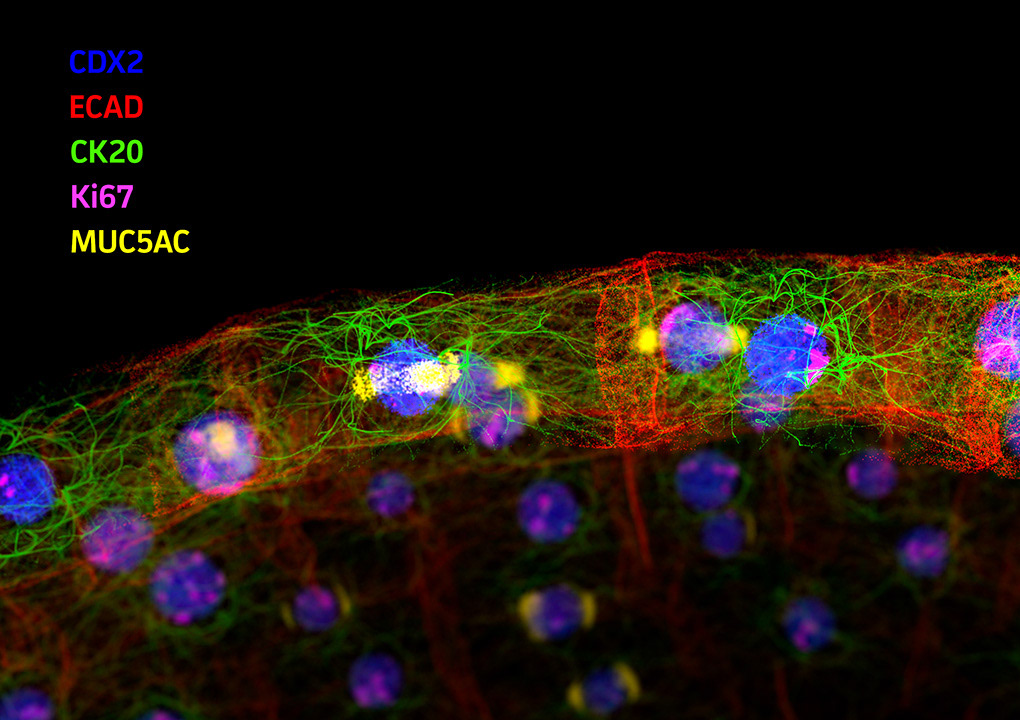
ATCC engaged Angstrom Images to produce an introductory animation for their Organoid product lines. Organoids – complex, self-organizing microtissues grown within an extracellular matrix – are proving to be invaluable models for the study of development and disease states. ATCC supplies organoids derived from a wide range of healthy and diseased tissues or cell lines, so the animation needed to be somewhat generic, while still depicting specific research uses for the model system.
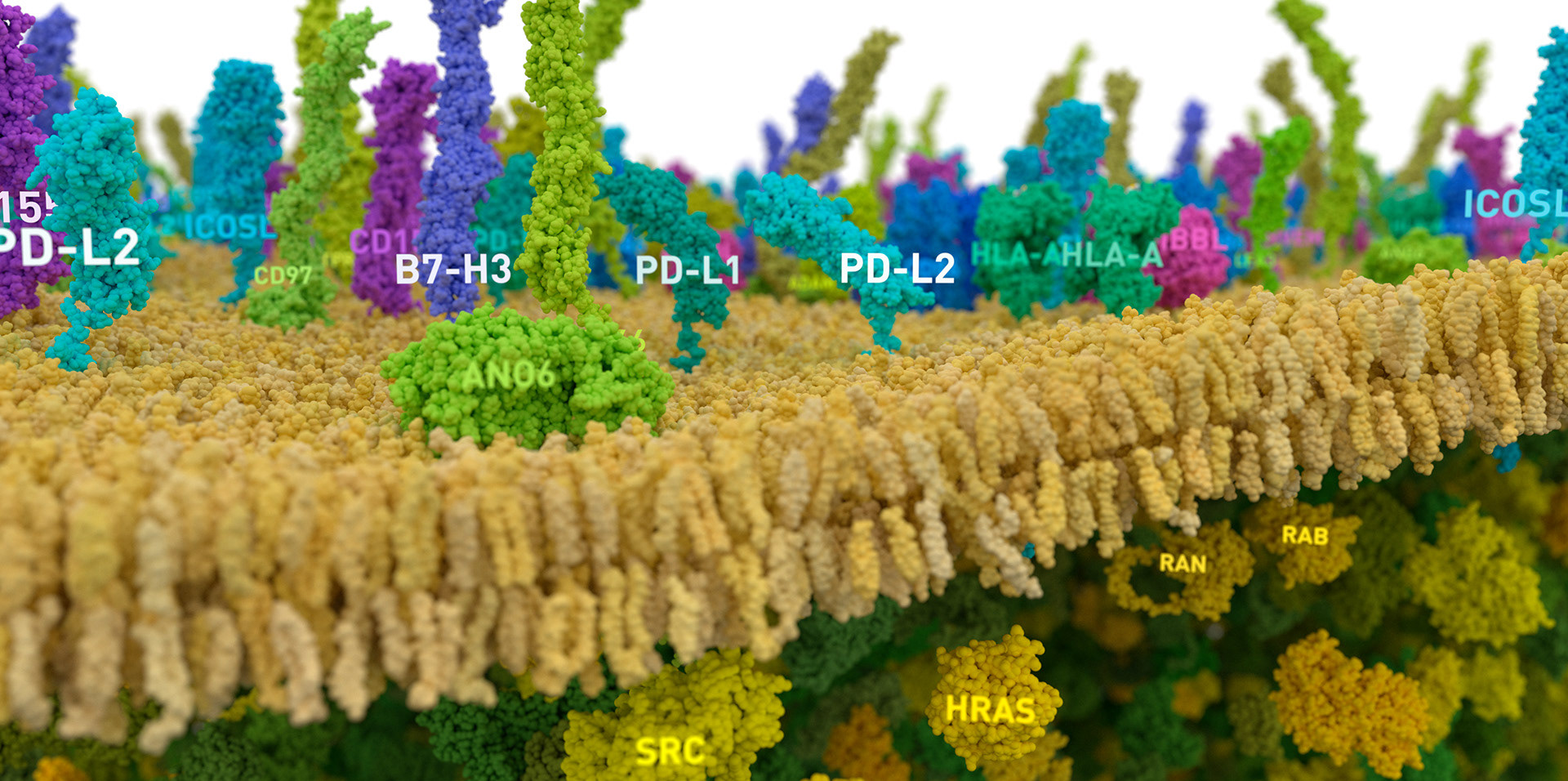
In this animation we show an early-stage organoid in a stylized ECM environment; cross-section to reveal typical internal structure; then apply a simulated immunofluorescence look to reveal tissue-level biomarker diversity. To close, we dive into a patch of membrane and show molecules relevant to cancer biology.

Technical Credits
Animations created by Evan Ingersoll at Angstrom Images, LLC, using SideFX Houdini, Maxon Redshift3d, and Adobe After Effects. Protein labels wrangled with Toadstorm’s MOPS tools for Houdini. All animation and still images Copyright ©2022 ATCC Corporation; used with permission.
Lipid geometry for cell membrane derived from supplemental materials from the LipidWrapper project; Durrant, Jacob & Amaro, Rommie. (2014). LipidWrapper: An Algorithm for Generating Large-Scale Membrane Models of Arbitrary Geometry. PLoS computational biology. 10. e1003720. DOI:10.1371/journal.pcbi.1003720.
The background protein population of the cell membrane and interior is based on proteomics data from:
Itzhak, Daniel N and Tyanova, Stefka and Cox, Jürgen and Borner, Georg HH. (2016) Global, quantitative and dynamic mapping of protein subcellular localization. eLife 2016;5:e16950
The Human Protein Atlas, proteinatlas.org; Thul PJ et al., A subcellular map of the human proteome. Science. (2017) PubMed: 28495876 DOI: 10.1126/science.aal3321
Itzhak, Daniel N and Tyanova, Stefka and Cox, Jürgen and Borner, Georg HH. (2016) Global, quantitative and dynamic mapping of protein subcellular localization. eLife 2016;5:e16950
The Human Protein Atlas, proteinatlas.org; Thul PJ et al., A subcellular map of the human proteome. Science. (2017) PubMed: 28495876 DOI: 10.1126/science.aal3321
Protein structures are derived from data in the RCSB PDB archive and the AlphaFold project:
rcsb.org, H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne. (2000) The Protein Data Bank Nucleic Acids Research, 28: 235-242.
Alphafold.ebi.ac.uk, Jumper, J et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021). Varadi, M et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research (2021).
AlphaFold data is used under a CC-BY-4.0 license.
rcsb.org, H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne. (2000) The Protein Data Bank Nucleic Acids Research, 28: 235-242.
Alphafold.ebi.ac.uk, Jumper, J et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021). Varadi, M et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research (2021).
AlphaFold data is used under a CC-BY-4.0 license.
For this project, protein structural data has been modified by
- Removing solvent, study compounds, and most ions
- Aligning related structures by semi-automated or manual processes
- Truncating propeptides
- Excising low-confidence regions of AlphaFold models
Molecular motion is simulated in Houdini, and does not represent realistic molecular dynamics. Membrane lipid composition and density is consistent with average values present in LipidWrapper data. Protein density was reduced for label legibility and collision avoidance, and is far below real-world values.
- Removing solvent, study compounds, and most ions
- Aligning related structures by semi-automated or manual processes
- Truncating propeptides
- Excising low-confidence regions of AlphaFold models
Molecular motion is simulated in Houdini, and does not represent realistic molecular dynamics. Membrane lipid composition and density is consistent with average values present in LipidWrapper data. Protein density was reduced for label legibility and collision avoidance, and is far below real-world values.